Shortly after their first large-scale computational annotation of long non-coding RNA (lncRNA) functions, which was published in the journal Nucleic Acids Research in May 2011, the Bioinformatics Research Group at the Institute of Computing Technology, Chinese Academy of Sciences, further released the first web service, ncFANs (non-coding RNA Function ANnotation server), to provide functional prediction of lncRNAs in human and mouse.
During the past few years, a large number of lncRNAs have been identified in mammalian transcriptome, particularly in human and mouse, which was facilitated by the rapid progress of next-generation sequencing technology. Unfortunately, however, most of them have not been functionally characterized due to the lack of relevant laboratory data. Therefore, excavating potential functions of lncRNAs presents an urgent challenge to the scientists in the field of bioinformatics.
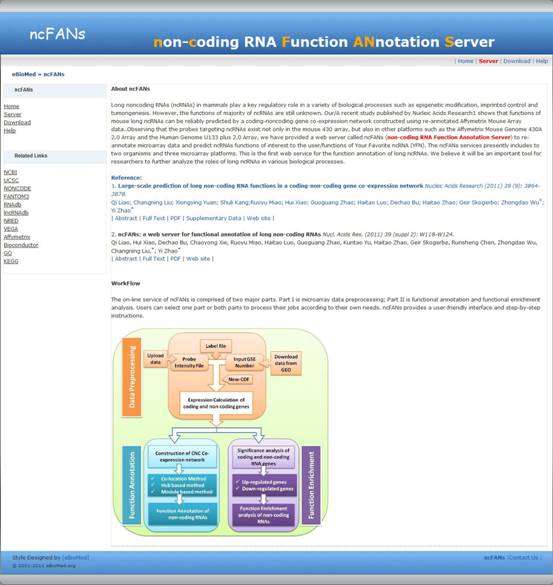
In their previous study, the group re-annotated the expression data from Affymetrix GeneChip Mouse Genome 430 2.0 Array, including both protein-coding genes and lncRNAs, and constructed a coding-non-coding gene co-expression (CNC) network. Based on topological or other network characteristics, such as module sharing, association with network hubs and combinations of co-expression and genomic adjacency, a total of 340 lncRNAs were able to be functionally annotated, which mainly involves organ or tissue development, cellular transport or metabolic processes.
The group’s current work has not only extended to the data in human but also developed a practical and user-friendly web interface for researchers to explore the functional annotation of human and mouse lncRNAs. On the basis of the re-annotated Affymetrix microarray data, ncFANs provides two alternative strategies for lncRNA functional annotation: one utilizes three aspects of the CNC network, whereas the other identifies condition-related differentially-expressed lncRNAs. The present version of ncFANs includes the re-annotated data from 10 human/mouse Affymetrix microarrays, and it will be continuously updated along with more lncRNA data from re-annotated microarray platforms as well as other high-throughput sources.
ncFANs has introduced a highly efficient way of re-using the abundant pre-existing microarray data. It is expected that the strategies developed in these studies will provide new clues and approaches for the study of lncRNA functions, and the predicted functions will give directions for molecular biological experiments.
ncFANs was published in the Web Server issue of Nucleic Acids Research in July 2011. It is freely accessible at http://www.ebiomed.org/ncFANs/ or http://www.noncode.org/ncFANs/.