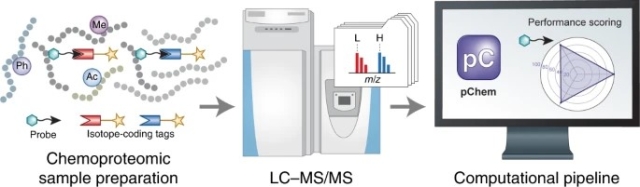
A new software tool, pChem, has been developed specifically for the detailed characterization of chemical probes in proteome-scale mass-spectrometry studies. The tool provides a comprehensive analysis of mass spectrometry data from chemoproteomics experiments.
With the development of tandem mass spectrometry-based protein identification methods, and in particular open search technology, the intersection application of high-throughput proteomics and other fields has expanded. Chemical proteomics established the bridge between chemical reaction research and protein sequence analysis, a combination helpful for characterizing drug-target interactions and for optimizing candidate drug molecules. At core, chemical proteomics offers breakthroughs in the design and synthesis of small-molecule chemical probes. A vital yet under-explored problem occurs with identifying and evaluating chemical probes’ characteristics when using experimental mass spectrometry data. The paper, “A modification-centric assessment tool for the performance of chemoproteomic probes,” published in Nature Chemical Biology, presented the first tool to help identify unknown modifications related to chemical probe reactions and to explore various applications.
pChem: A tool that assesses chemoproteomic probe performance
After summarizing current progress in open search engine technology based on mass spectrometry, and after analyzing advantages/disadvantages of existing open search algorithms, the researchers introduced pChem. This bioinformatic tool automatically determines the unknown mass modification and evaluates relevant properties. The pChem software combines open search and restricted search to determine the accurate mass and amino acid position selectivity of the target unknown modification. It will analyze the possible neutral loss of the unknown modification and fully describing its characteristics. The two-stage search framework effectively guarantees identification validity and results’ robustness. Corresponding statistical metrics reflect confidence in results for the reported identifications. To meet chemical probe design and application requirements, evaluation indicators were designed from three perspectives--profiling efficiency, modification homogeneity and residue selectivity. For convenience, users are able to intuitively estimate an experimental chemical probe’s performance.
The performance of the pChem tool was evaluated with an annotated standard chemical probe dataset. First, the researchers conducted an error analysis between the identification results of different strategies in pChem and the ground-truth target, which demonstrated the superiority of the current strategy selection. Second, identification results were compared with popular software MSFragger to demonstrate pChem software’s validity and the results’ robustness. According to test results on 18 public chemical probe data sets, our tool achieved a median error of 1.53 ppm in the target unknown modification mass, while accurate mass identification effectively reduced the number of candidates of inferred molecular formula.
By its extension in application research, the identification and evaluation of chemical probes offers more precise analysis of general unknown modifications in the blind search process. Its feedback can optimize the analysis and understanding of unknown modifications in common scenarios.
downloadFile